The concept
Automated control of synthetic microbial communities:
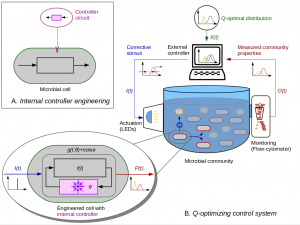 Engineering of biochemical circuits ensuring controlled single-cell response to actuation inputs
Engineering of biochemical circuits ensuring controlled single-cell response to actuation inputs
Mathematical modelling of dynamics of cellluar sub-populations and community interactions
Definition of community-level optimal control goals
Model-based estimation and optimal control algorithms
Computer-based automated control of bioreactor system
Achievement highlights
- Design, synthesis, characterization and control of a light-tunable differentiation system for bioproduction in yeast
- Design, synthesis and characterization of a consortium of E. coli bacteria for cooperative protein production
- New methods and results on the mathematical and numerical analysis of the microbial communities
- Design and theoretical analysis of real-time estimation and control methods for stabilization of biomasses/bioproduction performance
- Deployment of three platforms for the automated execution of monitoring and control experiments with microbial communities
Publications
In preparation/submitted:
- C. Martinez, E. Cinquemani, H. de Jong, J.-L. Gouzé, “Coexistence and optimization of a synthetic microbial community”
- A. dos Reis de Souza, D. Efimov, A. Polyakov, J.-L. Gouzé, E. Cinquemani, “State observation in microbial consortia: a case study on a synthetic producer-cleaner consortium”
- A. Pavlou, E.Cinquemani, J.Geiselmann, H. de Jong, “Protein-specific maturation models are necessary to obtain unbiased estimates of promoter activity”
In press:
- A. dos Reis de Souza, D. Efimov, T. Raïssi, X. Ping, “Robust output feedback model predictive control for constrained linear systems via interval observers”, Automatica
- A. Yabo, J.-B. Caillau, J.-L. Gouzé, H. de Jong, F. Mairet, “Dynamical analysis and optimization of a generalized resource allocation model of microbial growth”, SIADS.
https://hal.inria.fr/hal-03251044/file/maintenance.pdf - A. Yabo, J.-B. Caillau, J.-L. Gouzé, “Hierarchical MPC applied to bacterial resource allocation and metabolite synthesis”, Proceedings of CDC 2021. https://hal.archives-ouvertes.fr/hal-03189960/file/CDC2021.pdf
2021:
- A. Marguet, E. Cinquemani, “Identification of stochastic gene expression models over lineage trees”, Proceedings of the 19th IFAC sympusium on System Identification (SysId), Padova/online, July 13-16
- D. Lunz, G. Batt, J. Ruess, J. F. Bonnans, “Beyond the chemical master equation: stochastic chemical kinetics coupled with auxiliary processes”, PLoS Computational Biology. https://doi.org/10.1371/journal.pcbi.1009214
- D. Lunz, “On rapid oscillations driving biological processes at disparate timescales”, Physical Biology, 18(3):036002
- D. Lunz, “On continuum approximations of discrete-state Markov processes of large system size”, Multiscale Modeling and Simulation, SIAM, 19(1):294-319
- Z. R. Fox, S. Fletcher, A. Fraisse, C. Aditya, S. Sosa-Carrillo, S. Gilles, F. Bertaux, J. Ruess, G. Batt, “MicroMator: Open and Flexible Software for Reactive Microscopy”, bioRxiv. https://doi.org/10.1101/2021.03.12.435206
- C. Aditya, F. Bertaux, G. Batt, J. Ruess, “A light tunable differentiation system for the creation and control of consortia in yeast”, Nature Communications, 12:5829.
- F.Bertaux, J.Ruess, G.Batt, “External control of microbial populations for bioproduction: A modeling and optimization viewpoint”. Current Opinion in Systems Biology, 28:100394
- C.Aditya, F.Bertaux, G.Batt, J.Ruess, “Using single-cell models to predict the functionality of synthetic circuits at the population scale”, bioRxiv 2021.08.03.454887. https://doi.org/10.1101/2021.08.03.454887
- C. Martínez, J.-L. Gouzé, “Global dynamics of the chemostat with overflow metabolism”, Journal of Mathematical Biology, 82:3
- A. dos Reis de Souza, D. Efimov, T. Raïssi, “Robust output feedback MPC for LPV systems using interval observers”, IEEE Transactions on Automatic Control
- D. Lunz, G. Batt, J. Ruess, “To quarantine, or not to quarantine: A theoretical framework for disease control via contact tracing”, Epidemics, 34:100428. https://doi.org/10.1016/j.epidem.2020.100428
2020:
- F. Bertaux, S. Sosa-Carrillo, A. Fraisse, C. Aditya, M. Furstenheim, G. Batt, “Enhancing bioreactor arrays for automated measurements and reactive control with ReacSight”, bioRxiv, https://doi.org/10.1101/2020.12.27.424467
- A. dos Reis de Souza, J.-L. Gouzé, D. Efimov, A. Polyakov, “Robust adaptive estimation in the competitive chemostat”, Computers and Chemical Engineering, 142:107030, 2020
- A.G. Yabo, J.-L. Gouzé, “Optimizing bacterial resource allocation: metabolite production in continuous bioreactors”, Proceedings of the 21th IFAC World Congress, 2020
- E. Cinquemani, “Inference of the statistics of a modulated promoter process from population snapshot gene expression data”, Proceedings of the 21th IFAC World Congress, 2020
- A. dos Reis de Souza, D. Efimov, A. Polyakov, J.-L. Gouzé, “Observer-Based Robust Control of a Continuous Bioreactor with Heterogeneous Community”, Proceedings of the 21th IFAC World Congress, 2020
- A.G. Yabo, J.-B. Caillau, J.-L. Gouzé, “Optimal bacterial resource allocation: metabolite production in continuous bioreactors”, Mathematical Biosciences and Engineering, 17(6): 7074-7100, 2020
- M. Mauri, J.-L. Gouzé, H. de Jong, E. Cinquemani, “Enhanced production of heterologous proteins by a synthetic microbial community: Conditions and trade-offs”, PLoS Computational Biology, 16(4):e1007795, 2020
- A. dos Reis de Souza, D. Efimov, A. Polyakov, J.-L. Gouzé, “Robust stabilization of competing species in the chemostat”, Journal of Process Control, 87:138-146, 2020
2019:
- A. Yabo, J.-B. Caillau, J.-L. Gouzé, “Singular regimes for the maximization of metabolite production”, Proceedings of the 58th IEEE Conference on Decision and Control (CDC), 2019
- A. dos Reis de Souza, D. Efimov, A. Polyakov, J.-L. Gouzé, “On adaptive estimation of bacterial growth in the competitive chemostat”, Proceedings of the 11th IFAC Symposium on Nonlinear Systems Control (NOLCOS), 2019
- A. dos Reis de Souza, D. Efimov, A. Polyakov, J.-L. Gouzé, “Robust control of a competitive environment in the chemostat using discontinuous control laws”, Proceedings of the 58th IEEE Conference on Decision and Control (CDC), 2019
- A. Marguet, M. Lavielle, E. Cinquemani, “Inheritance and variability of kinetic gene expression parameters in microbial cells: Modelling and inference from lineage tree data”. Bioinformatics (Proceedings of ISMB/ECCB), 35(14):i586–i595, 2019
- E. Cinquemani, F. Mairet, I. Yegorov, H. de Jong, J.-L. Gouzé, “Optimal control of bacterial growth for metabolite production: The role of timing and costs of control”, Proceedings of the 17th European Control Conference (ECC), 2019
- E. Weill, V. Andreani, C. Aditya, P. Martinon, J. Ruess, G. Batt, J.F. Bonnans, “Optimal control of an artificial microbial differentiation system for protein bioproduction”, Proceedings of the 17th European Control Conference (ECC), 2019
- E. Cinquemani, “Stochastic reaction networks with input processes: Analysis and application to gene expression inference”, Automatica, 101:150-156, 2019
2018:
- E. Cinquemani, “Identifiability and Reconstruction of Biochemical Reaction Networks from Population Snapshot Data”, Processes (Special Issue on Computational Synthetic Biology), 6(9):136, 2018
