Introduction
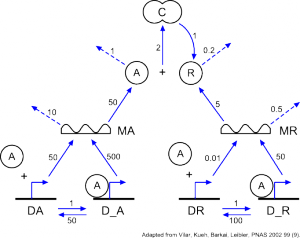
To measure the performance of PLASMA’s simulation engine applied to systems biology, we constructed plausible biological models of increasing complexity using the genetic oscillator of (Vilar, Kueh, Barkai, Leibler 2002) as a building block.
Algorithm
Here is the genosc1 model. This model is in the Reactive Module Languages.
Important!
// Based on genetic oscillator of Vilar, Kueh, Barkai, Leibler PNAS 2002
ctmc
const int MAX_VALUE = 10000;
const int alpha_A = 50;
const int Alpha_A = 500;
const double alpha_R = 0.01;
const int Alpha_R = 50;
const int beta_A = 50;
const int beta_R = 5;
const int delta_MA = 10;
const double delta_MR = 0.5;
const int delta_A = 1;
const double delta_R = 0.2;
const int gamma_A = 1;
const int gamma_R = 1;
const int gamma_C = 2;
const int theta_A = 50;
const int theta_R = 100;
module genosc1
DA : [0..MAX_VALUE] init 1;
D_A : [0..MAX_VALUE] init 0;
DR : [0..MAX_VALUE] init 1;
D_R : [0..MAX_VALUE] init 0;
MA : [0..MAX_VALUE] init 0;
MR : [0..MAX_VALUE] init 0;
A : [0..MAX_VALUE] init 0;
R : [0..MAX_VALUE] init 0;
C : [0..MAX_VALUE] init 0;
[] true -> DA*alpha_A : (MA’=MA+1);
[] true -> A*DA*gamma_A : (DA’=DA-1) & (D_A’=D_A+1) & (A’=A-1);
[] true -> D_A*Alpha_A : (MA’=MA+1);
[] true -> DR*alpha_R : (MR’=MR+1);
[] true -> D_R*Alpha_R : (MR’=MR+1);
[] true -> MA*beta_A : (A’=A+1);
[] true -> MR*beta_R : (R’=R+1);
[] true -> A*R*gamma_C : (A’=A-1) & (R’=R-1) & (C’=C+1);
[] true -> C*delta_A : (R’=R+1) & (C’=C-1);
[] true -> A*delta_A : (A’=A-1);
[] true -> R*delta_R : (R’=R-1);
[] true -> MA*delta_MA : (MA’=MA-1);
[] true -> MR*delta_MR : (MR’=MR-1);
[] true -> D_R*theta_R : (DR’=DR+1) & (D_R’=D_R-1) & (A’=A+1);
[] true -> A*DR*gamma_R : (DR’=DR-1) & (D_R’=D_R+1) & (A’=A-1);
[] true -> D_A*theta_A : (DA’=DA+1) & (D_A’=D_A-1) & (A’=A+1);
endmodule
Here is our model in Biological language.
Important!
constant alphaA=50,alpha_A=500,alphaR=0.01,alpha_R=50,betaA=50,betaR=5 // enzymatic
constant gammaA=1,gammaR=1,gammaC=2 // bimolecular
constant deltaA=1,deltaR=0.2,deltaMA=10,deltaMR=0.5,thetaA=50,thetaR=100// decay
species DA=1,D_A,DR=1,D_R,MA,MR,A,R,C
A + DA gammaA-> D_A
D_A thetaA-> A + DA
A + DR gammaR-> D_R
D_R thetaR-> A + DR
D_A alpha_A-> MA + D_A
DA alphaA-> MA + DA
D_R alpha_R-> MR + D_R
DR alphaR-> MR + DR
MA betaA-> MA + A
MR betaR-> MR + R
A + R gammaC-> C
C //deltaA
0.5-> R
A deltaA-> *
R deltaR-> *
MA deltaMA-> *
MR deltaMR-> *
//plot A,R,C
Download : genosc.plasma