Introduction
Treerecs allows for the correction of one or more gene trees with respect to a reference species tree. These trees are thus required as inputs.
In addition, the leaves from the gene tree(s) must be associated to their corresponding species in the species tree. This can be achieved in different ways which we will walk you through further on.
Required input:
- A reference species tree
- One or more gene tree(s) to be corrected
Ouput:
- One or more corrected gene tree(s) for each input gene tree
A simple example
You will find a very simple example in the examples/tutorial/1-simple directory
Input
Let’s go into this directory and checkout its content:
$ cd path/to/treerecs/examples/tutorial/1-simple
$ ls
gene_tree species_tree
$ cat species_tree
((a, b), c);
$ cat gene_tree
(a1:2.52, a2:2.15, b1:1.61, b2:1.93, b3:1.81, c1:3.4);
For those of you who are not familiar with the newick format, here’s a graphical representation of these trees:
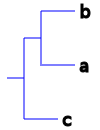
Species tree
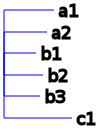
Gene tree
Run Treerecs
Let’s run the following command and check the result:
$ treerecs -s species_tree -g gene_tree
Treerecs, Inria - team Beagle, 2017
Solution saved in treerecs_output/gene_tree_recs.nwk
Total elapsed time 0.011 seconds.
Inspect output
$ cat treerecs_output/gene_tree_recs.nwk
> family 1 tree 1 (total cost = 4, duplications = 2, losses = 0, contraction threshold = 0, execution time = 0.003 s.)
(c1:1.7,((a2:2.15,b3:1.81):1e-06,(a1:2.52,(b1:1.61,b2:1.93):1e-06):1e-06):1.7);
Graphical representation:
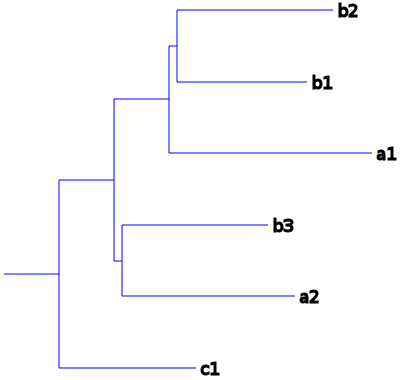
Corrected gene tree
Graphical output
Here, we have used an external newick visualization tool to generate the graphical representation of our graphs.
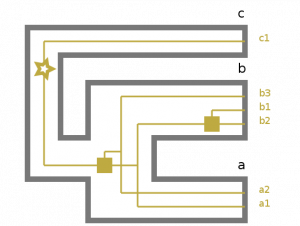
There is actually an option to get a graphical output straight from Treerecs. It outputs an SVG displaying the gene trees embedded in the species tree.
To get this output, use the -O switch with value svg:
$ treerecs -s species_tree -g gene_tree -O svg -o treerecs_output_with_svg
Treerecs, Inria - team Beagle, 2017
Solution saved in treerecs_output_with_svg/gene_tree_recs.svg
Total elapsed time 0.01 seconds.
And here’s the generated SVG (the star represents the gene tree root, squares represent gene duplications):
Specifying the gene-species mapping
When no explicit mapping is needed
In the simple example above, we did not have to worry about the gene-species mapping, i.e. about specifying which species each gene corresponds to. This is because all the genes in the gene tree had the corresponding species name embedded in their own name. For example, gene a1 corresponds to species a (“a” is a substring of “a1”). Usually, this is more explicit: for example, gene ENSP00000364946 belonging to species Homo-sapiens can be named Homo-sapiens_ENSP00000364946.
To make it short, whichever format is used for the gene names, if for each gene the corresponding species name is a substring of the gene name, Treerecs can build the gene-species mapping automatically from this information.
Alternatively, Treerecs can use the NHX ‘S’ (species name) tag to build the mapping.
Explicit mapping
If your data does not allow for automatic mapping, you will have to provide the gene-species mapping explicitly as a separate file.
You will find an example in the examples/tutorial/2-mapping directory
If you try running Treerecs as we did before with only the gene tree and species tree as input, Treerecs will complain it can’t map some genes to any species:
$ cd path/to/treerecs/examples/tutorial/2-mapping
$ treerecs -s species_tree -g gene_tree
Treerecs, Inria - team Beagle, 2017
Error during gene <> species mapping, some gene leaves cannot be mapped:
a1, a2.
Indeed, in this example, the species names are aaa, b and c. So genes a1 and a2 don’t include a species name in their own name and thus can’t be mapped automatically.
Treerecs can be used in such a setting by handing it a mapping file. This file must contain one line per gene, each line consisting of the gene name, a white space and the name of the corresponding species.
For example, in our example, the mapping file contains:
a1 aaa
a2 aaa
b1 b
b2 b
b3 b
c1 c
You can then run Treerecs with the -S option as follows:
$ treerecs -s species_tree -g gene_tree -S mapping
Treerecs, Inria - team Beagle, 2017
Solution saved in treerecs_output/gene_tree_recs.svg
Total elapsed time 0.01 seconds.

